Mollusk-omics
Mollusk-omics
Molluscan comparative genomics
One of our main goals is to understand how the great diversity of body plans present in modern mollusks arose through evolutionary processes from a common ancestral species. In addition to our morhoevodevo work, we utilize comparative genomic and transcriptomic approaches to gain insight into the genetic developments that have underpinned lineage specific innovations. We investigate protein coding, non-coding and regulatory genetic evolution in mollusks with a strong focus on developmental transcriptional regulation.
Ongoing:
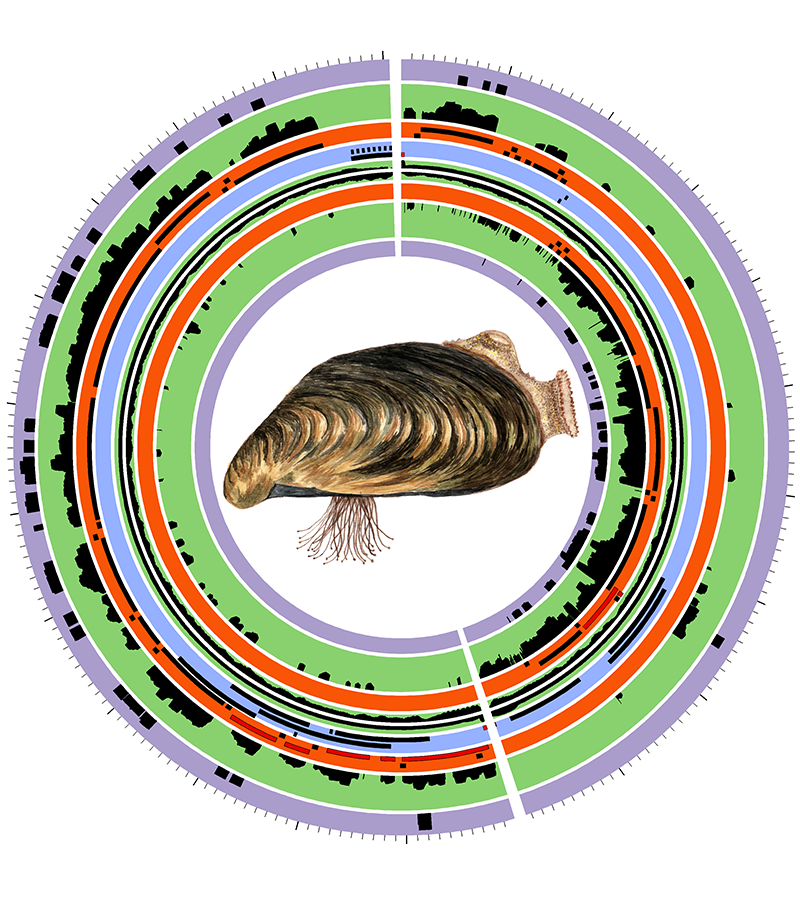
> Genome assembly and annotation of the chiton Acanthochitona fascicularis (Calcino)
> Developmental transcriptomics in molluscan mitochondria (Calcino)
> Co-expressed gene networks and core promoter characterization in the quagga mussel (Calcino)
> Long non-coding RNAs in mollusks (Calcino)
Recently completed:
> The quagga mussel genome and the evolution of freshwater tolerance (Calcino)
> Ancient origins of arthropod molting pathway components (De Oliveira)
> Extensive conservation of the proneuropeptide and peptide hormone complement in mollusks (De Oliveira)
Single cell transcriptomics
Single cell transcriptomics is providing us with information on unique transcriptional profiles of individual cell types. This transformative technology is a powerful tool for determining the homology of cell types between species, for tracking the genetic changes that occur during cell type differentiation and for isolating genes involved in specific genetic pathways. Single cell transcriptomics has particular utility during embryogenesis when specific cell types or tissues cannot easily be dissected or isolated. We anticipate that this technique will become increasingly important to our work understanding molluscan developmental processes.
Ongoing:
> Single cell sequencing in the invasive quagga mussel (Salamanca)

Development of tools for genetic manipulation
The comprehensive insights in to developmental gene regulation obtained through our comparative genomics approaches have provided a backbone on which we can now more thoroughly investigate gene function through innovative manipulative techniques. We are beginning to develop gene-editing protocols using the CRISPR/Cas9 system for gene knockout, in addition to GFP reporter plasmid protocols to visualize developmental gene expression dynamics. As the largest phylum of the Lophotrochozoa, mollusks are uniquely situated to inform on the hypothesized ancestral functions of genes which have traditionally only been investigated in a small number of model ecdysozoans (e.g. insects and nematodes) and deuterostomes (e.g. vertebrates).
Ongoing:
> GFP reporter plasmid design and delivery to developing quagga mussels (Dreissena rostriformis)
> CRISPR/Cas9 cassette design
Study & research opportunities
Interested in doing an internship, academic thesis or postdoc in mollusk-omics? Please contact Andrew Calcino or Andreas Wanninger.
